Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
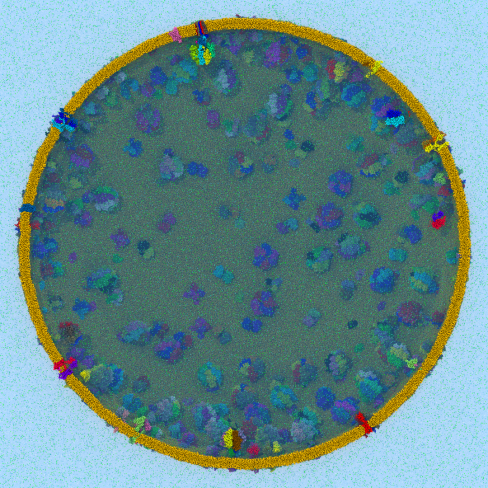
Disease due to a viral or bacterial infection continues to pose a major health concern, particularly today in the midst of the COVID-19 pandemic. Fighting such health threats requires the development of new antiviral and antibiotic drugs, a process that is increasingly guided by molecular modeling and computer simulation. The recently released NAMD 2.14 is now especially well suited to handle very large molecular systems for cell-scale simulations and to take advantage of massively parallel, GPU-accelerated architectures like the Summit supercomputer at Oak Ridge National Laboratory, currently ranked as the fastest in the world. Recent Summit simulations of a one-billion-atom protocell system, assembled to emulate a minimal but realistic cellular envelope and shown here in a cross-sectional view, push the limits of constructing and modeling atomic-resolution biological structures, paving the way for improved methodologies and tools that will enable the simulation of large viral and bacterial complexes.